Research
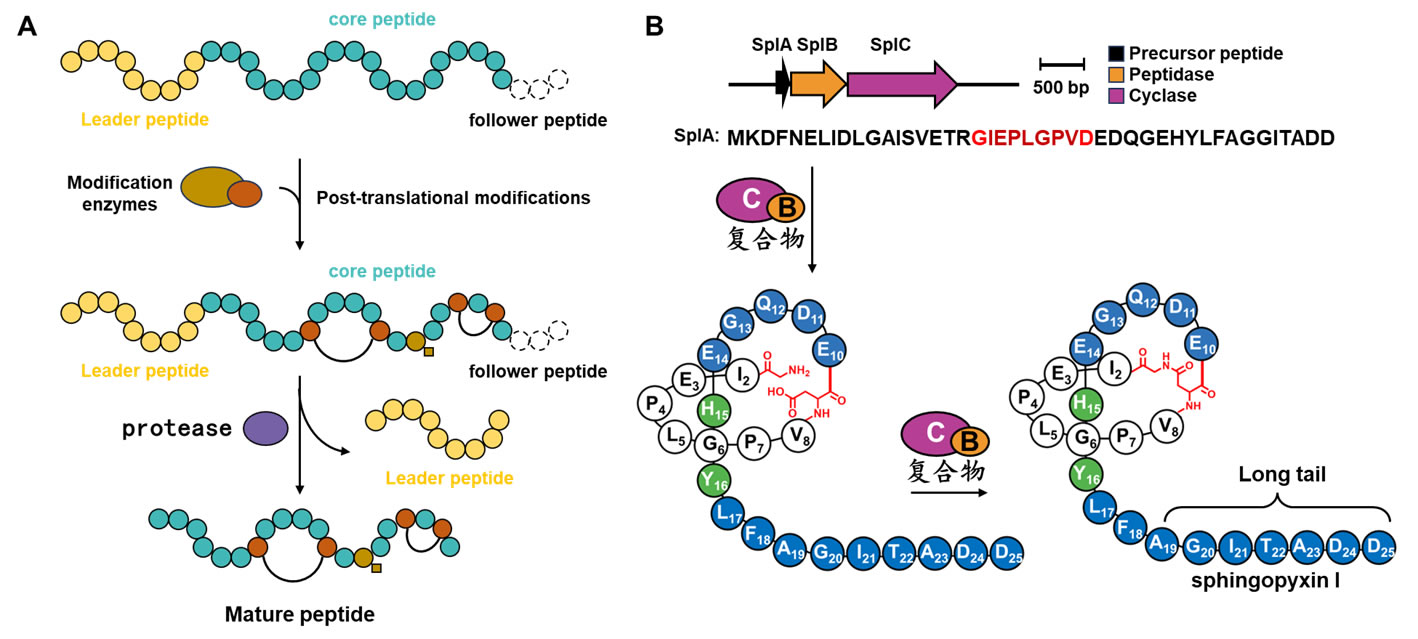
1. Study on the Biosynthesis Mechanisms of RiPPs
Ribosomally synthesized and post-translationally modified peptides (RiPPs) make up a rapidly growing superfamily of natural products. We employ a multidisciplinary approach, including biochemistry, structural biology, molecular biology, and chemistry, to investigate the biosynthetic mechanisms of complex peptide natural products with significant biological functions. Through bioinformatics methods, we identify complete biosynthetic gene clusters and establish fundamental biosynthetic pathways. Using X-ray crystallography and cryo-electron microscopy techniques, we explore the enzymatic mechanisms involved in the formation of unique structural units. Furthermore, we conduct genetic modifications to the biosynthetic pathways to expand the diversity of chemical structures and functionalities.
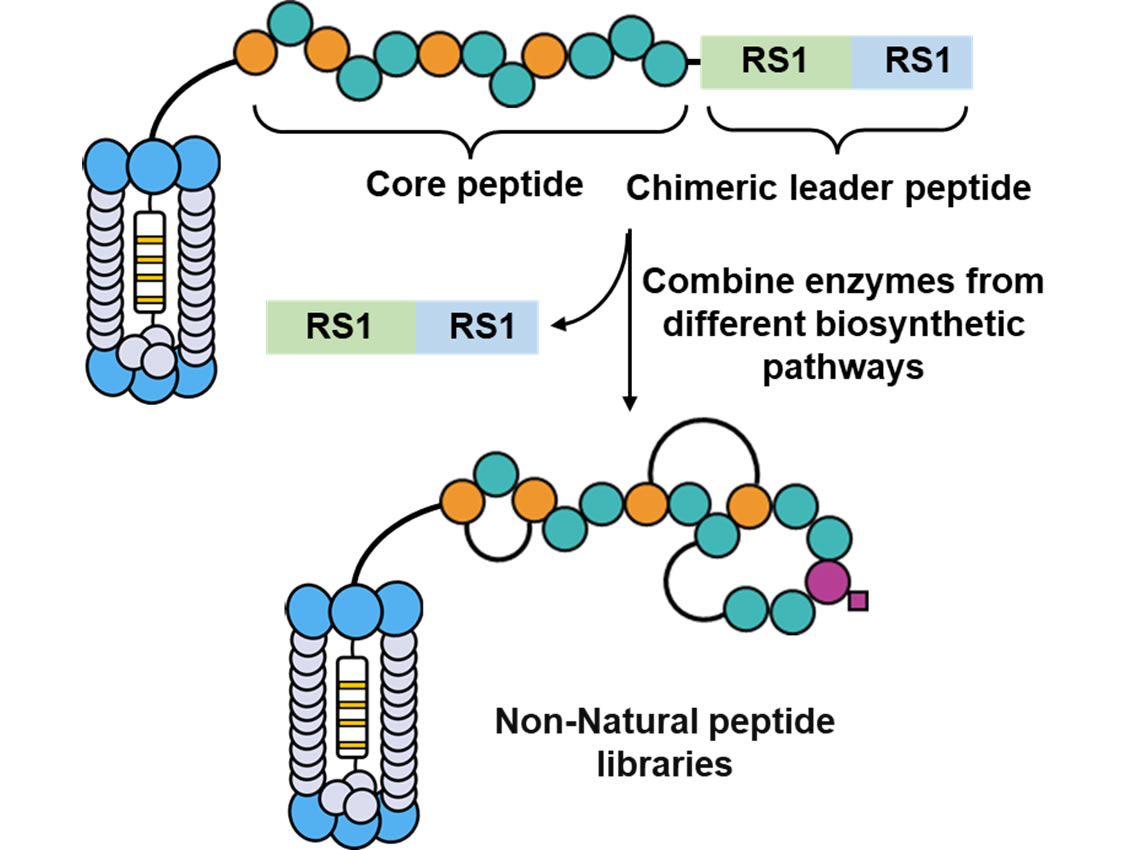
2. Phage Display Pseudo-natural Product Peptide Library
Since the pioneering work of George P. Smith in the development of phage display technology, the screening of peptide drug molecules has entered a new era. Many structurally novel peptide drugs have been successfully screened and applied in the treatment of diseases. However, phage display is limited to displaying peptide libraries composed of the 20 natural amino acids, which restricts its use in screening target proteins with complex structures. We aim to integrate the biosynthetic pathways of peptide natural products with phage display technology, thereby introducing various structurally complex non-natural building blocks into the phage-displayed peptide library of natural products, ultimately achieving a phage display library of natural product-like peptides.
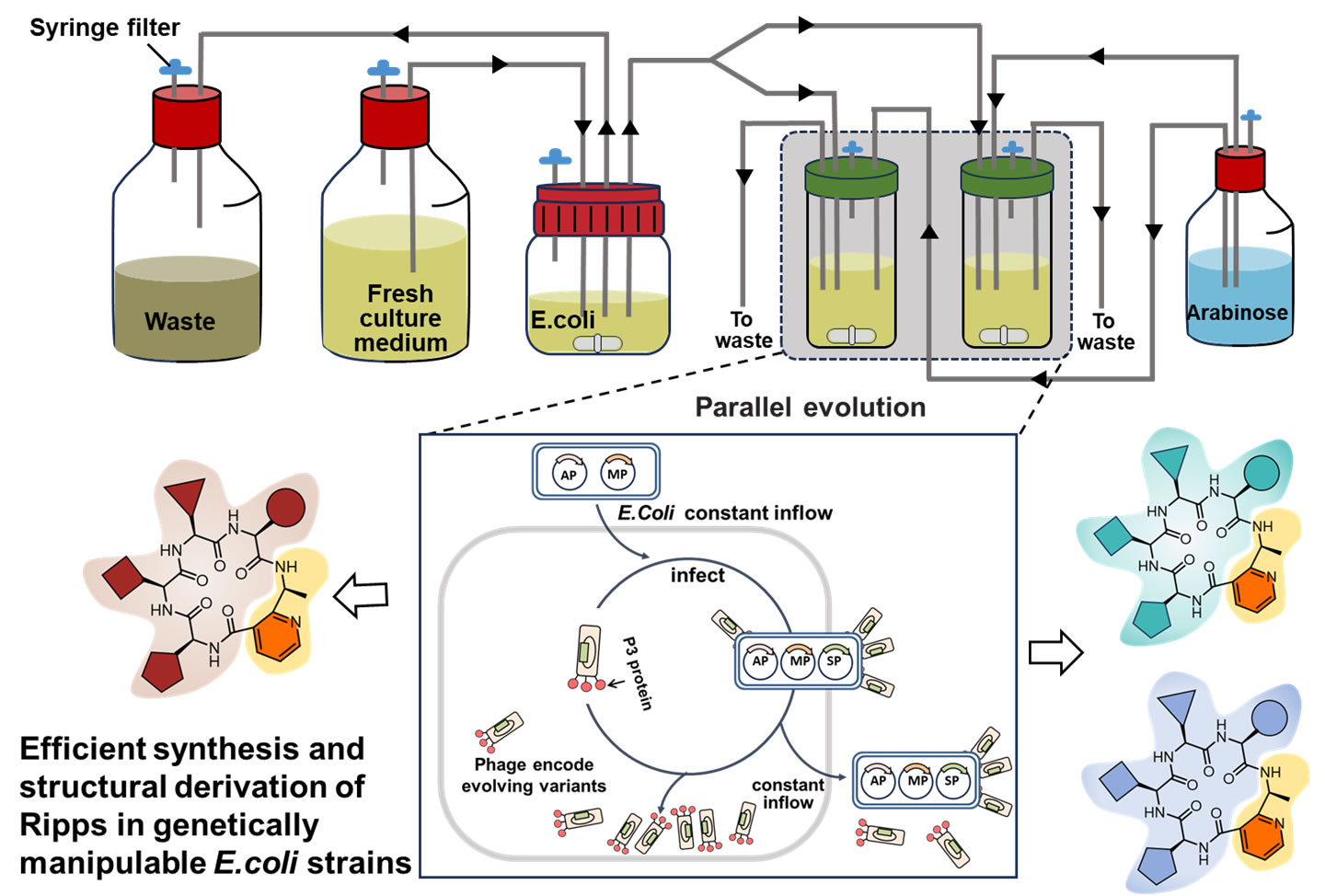
3. Phage-assisted Biosynthetic Enzyme Directed Evolution System
Peptide natural products often exhibit intricate structural diversity and possess significant biological activity, rendering them pivotal in the field of chemical biology and the development of innovative drugs throughout history. However, their complex structures pose challenges to current methods of chemical synthesis and biosynthesis. Based on the unique biosynthetic logic of ribosomal peptide natural products, we proposes an innovative approach that integrates phage life cycles with ribosomal peptide biosynthesis through ingenious design of gene transcription elements. Our goal is to establish a phage-assisted ribosomal peptide biosynthesis gene cluster directed evolution system, enabling the efficient synthesis and structural derivation of ribosomal peptides in genetically manipulable E.coli strains.